-Search query
-Search result
Showing all 49 items for (author: vargas & j)

EMDB-40059: 
Homo-octamer of PbuCsx28 protein
Method: single particle / : Park JU, Kellogg EH

PDB-8gi1: 
Homo-octamer of PbuCsx28 protein
Method: single particle / : Park JU, Kellogg EH

EMDB-15516: 
Cryo-EM Snapshots of Nanodisc-Embedded Native Eukaryotic Membrane Proteins
Method: single particle / : Janson K, Kyrilis FL, Tueting C, Alfes M, Das M, Traeger TK, Schmidt C, Hamdi F, Keller S, Meister A, Kastritis PL

EMDB-15517: 
myo-Inositol-1-Phosphate Synthase
Method: single particle / : Janson K, Kyrilis FL, Tueting C, Alfes M, Das M, Traeger TK, Schmidt C, Hamdi F, Keller S, Meister A, Kastritis PL
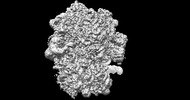
EMDB-26705: 
allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome
Method: single particle / : Zhang J, Krahn N, Prabhakar A, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J
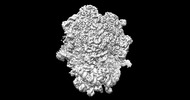
EMDB-26713: 
allo-tRNAUTu1A in the A site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J
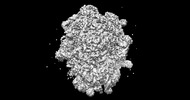
EMDB-26714: 
allo-tRNAUTu1A in the P site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

PDB-7ur5: 
allo-tRNAUTu1 in the A, P, and E sites of the E. coli ribosome
Method: single particle / : Zhang J, Krahn N, Prabhakar A, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

PDB-7uri: 
allo-tRNAUTu1A in the A site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

PDB-7urm: 
allo-tRNAUTu1A in the P site of the E. coli ribosome
Method: single particle / : Zhang J, Prabhakar A, Krahn N, Vargas-Rodriguez O, Krupkin M, Fu Z, Acosta-Reyes FJ, Ge X, Choi J, Crnkovic A, Ehrenberg M, Viani Puglisi E, Soll D, Puglisi J

EMDB-26322: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

PDB-7u32: 
MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution
Method: single particle / : Shan Z, Pye VE, Cherepanov P, Lyumkis D

EMDB-14453: 
MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution
Method: single particle / : Ballandras-Colas A, Nans A, Cherepanov P

EMDB-21558: 
30S-Activated-high-Mg2+
Method: single particle / : Jahagirdar D, Jha V

EMDB-21569: 
30S-Inactivated-high-Mg2+ Class A
Method: single particle / : Jahagirdar D, Jha V

EMDB-21570: 
30S-Inactive-high-Mg2+ Class B
Method: single particle / : Ortega J

EMDB-21571: 
30S-Inactive-high-Mg2+ + carbon layer
Method: single particle / : Jahagirdar D, Jha V

EMDB-21572: 
30S-Inactive-low-Mg2+ Class A
Method: single particle / : Jahagirdar D, Jha V

EMDB-21573: 
30S-Inactive-low-Mg2+ Class B
Method: single particle / : Jahagirdar D, Jha V

EMDB-22690: 
30S-Inactive-low-Mg2+ Carbon
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

PDB-6w6k: 
30S-Activated-high-Mg2+
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

PDB-6w77: 
30S-Inactivated-high-Mg2+ Class A
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

PDB-6w7m: 
30S-Inactive-high-Mg2+ + carbon layer
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

PDB-6w7n: 
30S-Inactive-low-Mg2+ Class A
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

PDB-6w7w: 
30S-Inactive-low-Mg2+ Class B
Method: single particle / : Jahagirdar D, Jha V, Basu B, Gomez-Blanco J, Vargas J, Ortega J

EMDB-20858: 
The inner junction complex of Chlamydomonas reinhardtii doublet microtubule
Method: single particle / : Khalifa AAZ, Ichikawa M, Bui KH

PDB-6ve7: 
The inner junction complex of Chlamydomonas reinhardtii doublet microtubule
Method: single particle / : Khalifa AAZ, Ichikawa M, Bui KH

EMDB-20855: 
48-nm repeat unit of the doublet microtubule from Chlamydomonas reinhardtii
Method: single particle / : Khalifa AAZ, Ichikawa M, Bui KH

EMDB-20856: 
16-nm repeat of the doublet microtubule from Tetrahymena thermophila
Method: single particle / : Ichikawa M, Khalifa AAZ, Bui KH

EMDB-20602: 
Cryo-EM structure of the 48-nm repeat unit of the doublet microtubule from Tetrahymena thermophila
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

EMDB-20603: 
Cryo-EM map of the A-tubule prepared by sonication
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

EMDB-20606: 
Cryo-EM map of the A-tubule prepared by 0.2% Sarkosyl treatment
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

PDB-6u0h: 
Tubulin lattice of the ciliary doublet microtubule from Tetrahymena thermophila
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

PDB-6u0t: 
Protofilament Ribbon Flagellar Proteins Rib43a-S
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

PDB-6u0u: 
Protofilament Ribbon Flagellar Proteins Rib43a-L
Method: single particle / : Ichikawa M, Khalifa AAZ, Vargas J, Basu K, Bui KH

EMDB-0481: 
Role of Era in Assembly and Homeostasis of the Ribosomal Small Subunit
Method: single particle / : Razi A, Davis JH, Hao Y, Jahagirdar D, Thurlow B, Basu K, Jain N, Gomez-Blanco J, Britton RA, Vargas J, Guarne A, Woodson SA, Williamson JR, Ortega J

EMDB-0482: 
Role of Era in Assembly and Homeostasis of the Ribosomal Small Subunit
Method: single particle / : Ortega J, Davis JH, Hao Y, Jahagirdar D, Thurlow B, Basu K, Jain N, Gomez-Blanco J, Britton RA, Vargas J, Guarne A, Woodson SA, Williamson JR

EMDB-0483: 
Role of Era in Assembly and Homeostasis of the Ribosomal Small Subunit
Method: single particle / : Ortega J, Davis JH, Hao Y, Jahagirdar D, Thurlow B, Basu K, Jain N, Gomez-Blanco J, Britton RA, Vargas J, Guarne A, Woodson SA, Williamson JR

EMDB-0484: 
Role of Era in Assembly and Homeostasis of the Ribosomal Small Subunit
Method: single particle / : Ortega J, Davis JH, Hao Y, Jahagirdar D, Thurlow B, Basu K, Jain N, Gomez-Blanco J, Britton RA, Vargas J, Guarne A, Woodson SA, Williamson JR

PDB-6nqb: 
Role of Era in Assembly and Homeostasis of the Ribosomal Small Subunit
Method: single particle / : Ortega J

EMDB-3825: 
Three-dimensional cryo-EM density map of paired C2S2M PSII-LHCII supercomplexes from thylakoid membranes of Pisum sativum
Method: single particle / : Pagliano C

EMDB-8718: 
Domain identification of Penicillium Chrysogenum UGGT by labeling the protein with monovalent streptavidin
Method: single particle / : Calles-Garcia D, Yang M, Vargas J, Melero R, Soya N, Menade M, Ito Y, Lukacs G, Kollman J, Kozlov G, Gehring K

EMDB-8719: 
The Electron Microscopy map of Drosophila melanogaster UDP-glucose: glycoprotein glucosyltransferase (UGGT)
Method: single particle / : Vargas J, Melero R, Yang M, Calles-Garcia D, Soya N, Menade M, Ito Y, Lukacs G, Kollman J, Kozlov G, Gehring K

EMDB-3540: 
Asymmetric reconstruction of MS2 virion
Method: single particle / : Koning RI, Gomez Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ

EMDB-8361: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

PDB-5t5h: 
Structure and assembly model for the Trypanosoma cruzi 60S ribosomal subunit
Method: single particle / : Liu Z, Gutierrez-Vargas C, Wei J, Grassucci RA, Ramesh M, Espina N, Sun M, Tutuncuoglu B, Madison-Antenucci S, Woolford Jr JL, Tong L, Frank J

EMDB-3402: 
Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ
Method: single particle / : Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ

EMDB-3403: 
Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ
Method: single particle / : Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ

EMDB-3404: 
Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ
Method: single particle / : Koning RI, Gomez-Blanco J, Akopjana I, Vargas J, Kazaks A, Tars K, Carazo JM, Koster AJ
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
